Keywords: Image processing, Information, Data analysis, Modeling, Supercomputing, Open source, Deep learning, Cell segmentation, Cell detection, Instance segmentation, Cell counting, Pytorch
Cell Detection
⭐ Showcase
NeurIPS 22 Cell Segmentation Competition
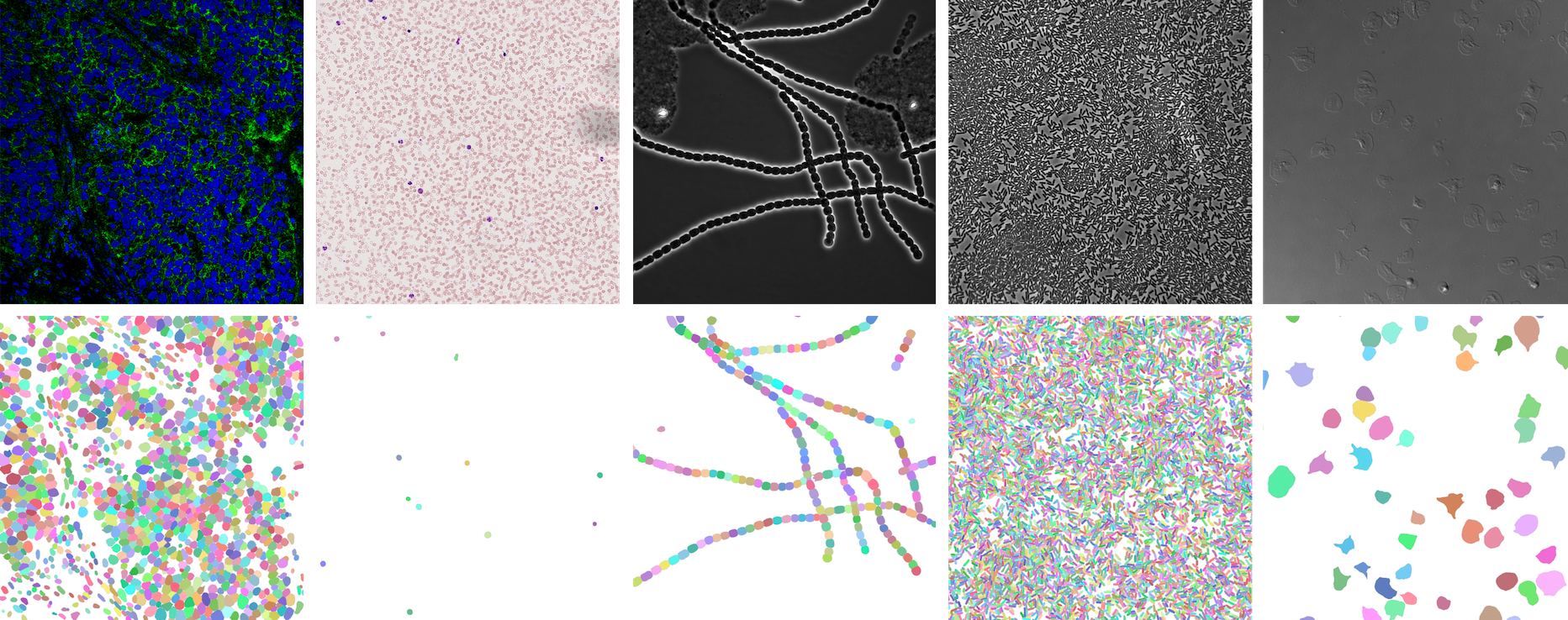
https://openreview.net/forum?id=YtgRjBw-7GJ
Nuclei of U2OS cells in a chemical screen
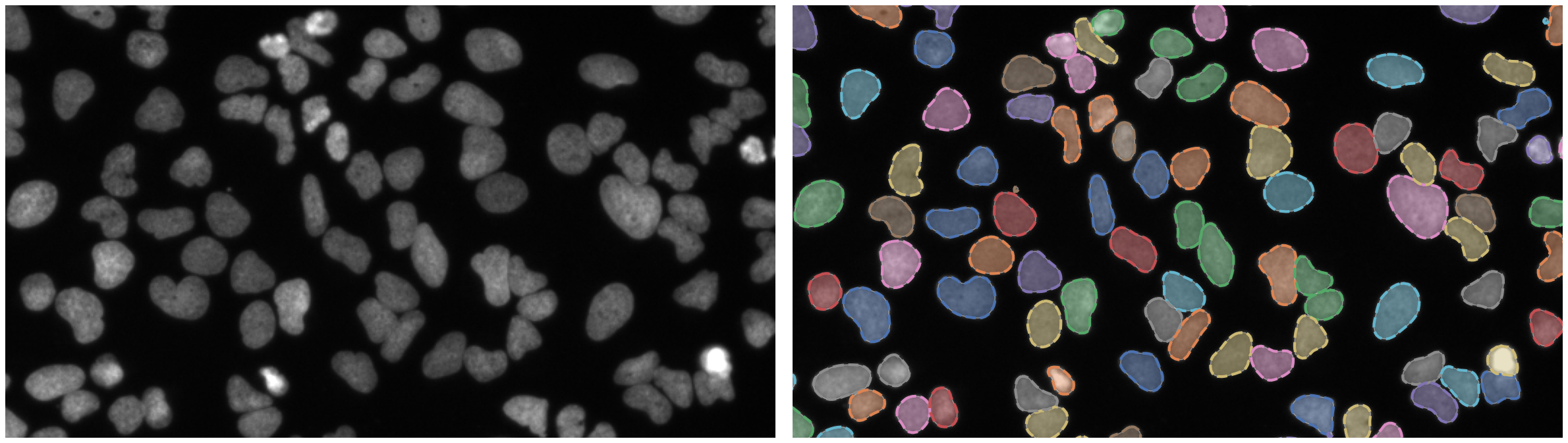
https://bbbc.broadinstitute.org/BBBC039 (CC0)
P. vivax (malaria) infected human blood
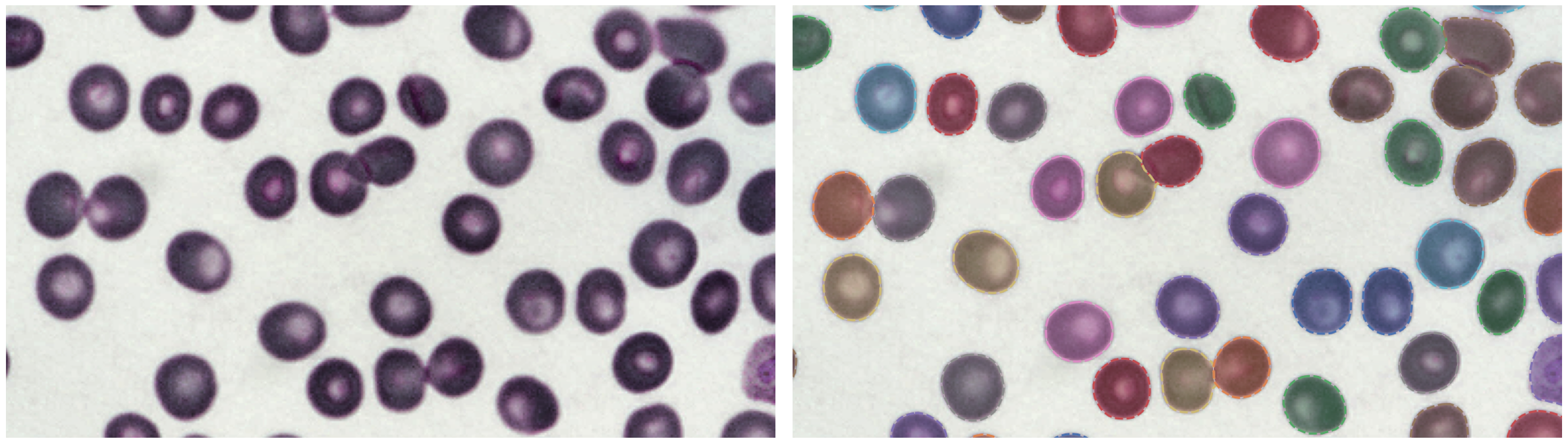
https://bbbc.broadinstitute.org/BBBC041 (CC BY-NC-SA 3.0)
🛠 Install
Make sure you have PyTorch installed.
PyPI
pip install -U celldetection
GitHub
pip install git+https://github.com/FZJ-INM1-BDA/celldetection.git
💾 Trained models
model = cd.fetch_model(model_name, check_hash=True)
| model name | training data | link |
|---|---|---|
ginoro_CpnResNeXt101UNet-fbe875f1a3e5ce2c |
BBBC039, BBBC038, Omnipose, Cellpose, Sartorius - Cell Instance Segmentation, Livecell, NeurIPS 22 CellSeg Challenge | 🔗 |
🐳 Docker
Find us on Docker Hub: https://hub.docker.com/r/ericup/celldetection
You can pull the latest version of celldetection via:
doker pull ericup/celldetection:latest
Apptainer
You can also pull our Docker images for the use with Apptainer (formerly Singularity) with this command:
apptainer pull --dir . --disable-cache docker://ericup/celldetection:latest
🤗 Hugging Face Spaces
Find us on Hugging Face and upload your own images for segmentation: https://huggingface.co/spaces/ericup/celldetection
🧑💻 Napari Plugin
Find our Napari Plugin here: https://github.com/FZJ-INM1-BDA/celldetection-napari
Find out more about Napari here: https://napari.org
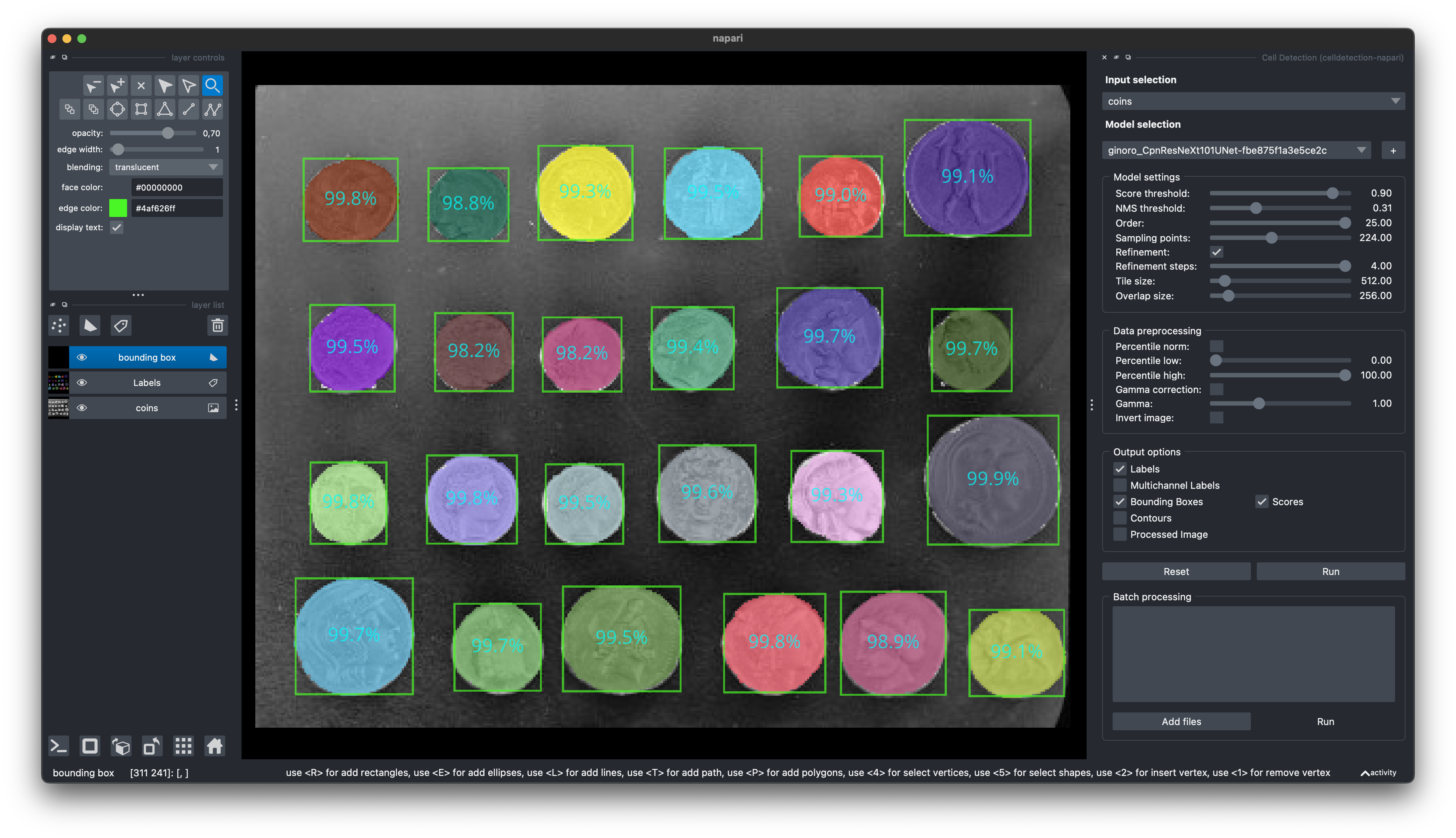
You can install it via pip:
pip install git+https://github.com/FZJ-INM1-BDA/celldetection-napari.git
🏆 Awards
- NeurIPS 2022 Cell Segmentation Challenge: Winner Finalist Award
📝 Citing
If you find this work useful, please consider giving a star ⭐️ and citation:
@article{UPSCHULTE2022102371,
title = {Contour proposal networks for biomedical instance segmentation},
journal = {Medical Image Analysis},
volume = {77},
pages = {102371},
year = {2022},
issn = {1361-8415},
doi = {https://doi.org/10.1016/j.media.2022.102371},
url = {https://www.sciencedirect.com/science/article/pii/S136184152200024X},
author = {Eric Upschulte and Stefan Harmeling and Katrin Amunts and Timo Dickscheid},
keywords = {Cell detection, Cell segmentation, Object detection, CPN},
}
🔗 Links
Publications
The multimodality cell segmentation challenge: toward universal solutions
Ma J, Xie R, Ayyadhury S, Ge C, Gupta A, Gupta R, Gu S, Zhang Y, Lee G, Kim J, Lou W, Li H, Upschulte E, Dickscheid T, de Almeida J, Wang Y, Han L, Yang X, Labagnara M, Gligorovski V, Scheder M, Rahi S, Kempster C, Pollitt A, Espinosa L, Mignot T, Middeke J, Eckardt J, Li W, Li Z, Cai X, Bai B, Greenwald N, Van Valen D, Weisbart E, Cimini B, Cheung T, Brück O, Bader G, Wang B - Nature Methods - 2024
CellDetection
Upschulte E, Harmeling S, Amunts K, Dickscheid T - Zenodo - 2023
Contour proposal networks for biomedical instance segmentation
Upschulte E, Harmeling S, Amunts K, Dickscheid T - Medical Image Analysis - 2022


